Hi all,
I’m trying to extract a set of tagged facets from a 2D mesh.
I’ve created the following script:
try:
import netgen.geom2d
except ModuleNotFoundError:
print("This example requires the netgen-mesher module to be installed.")
exit(1)
import numpy as np
geo = netgen.geom2d.CSG2d()
poly = netgen.geom2d.Solid2d(
[
(0, 0),
netgen.geom2d.EdgeInfo(bc="bottom"),
(2, 0),
netgen.geom2d.EdgeInfo(bc="right"),
(2, 2),
netgen.geom2d.EdgeInfo(bc="top"),
(0, 2),
netgen.geom2d.EdgeInfo(bc="left"),
]
)
geo.Add(poly)
ngmesh = geo.GenerateMesh(maxh=2)
ngmesh.Export("tmp.mesh", "Medit Format")
regions: dict[str : list[int]] = {}
for region in range(len(ngmesh.GetRegionNames(codim=1))):
if (name := ngmesh.GetBCName(region)) in regions.keys():
regions[name].append(region + 1)
else:
regions[name] = [region + 1]
print(f"{regions=}")
I then in turn try to create a relationship between he edge-info string, and the indices (integer marker of the edges).
I am not able to understand the relationship between them, as regions prints:
regions={'bottom': [1], 'right': [2], 'top': [3], 'left': [4]}
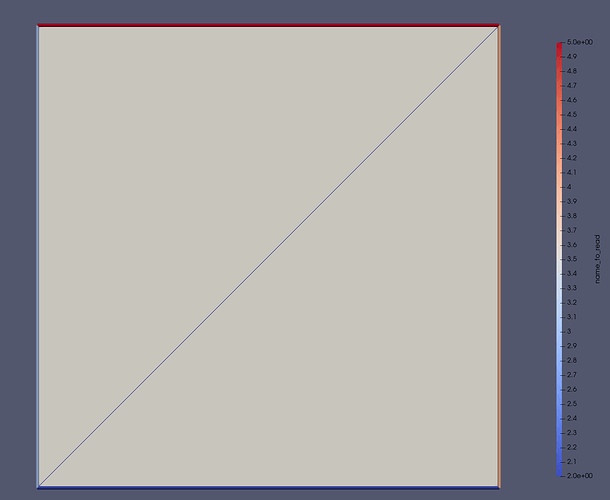
resulting in the mesh
Further inspection of the facets/edges yield
ng_facets = ngmesh.Elements1D()
facet_indices = ng_facets.NumPy()["nodes"].astype(np.int64)
facets = (
np.array(
[list(np.trim_zeros(a, "b")) for a in list(facet_indices)],
dtype=np.int64,
)
- 1
)
facet_values = ng_facets.NumPy()["index"].astype(np.int32)
print("Unique facet values:", np.unique(facet_values))
with output
Unique facet values: [1 2 3 4]
but then mapping the facets to its coordinates, reveal a mismatch:
x = ngmesh.Coordinates()
for region, indices in regions.items():
print(f"{region=}")
for index in indices:
facet_positions = np.flatnonzero(facet_values == index)
print(f"{x[facets[facet_positions]]=}")
returns
region='bottom'
x[facets[facet_positions]]=array([[[0., 0.],
[2., 0.]]])
region='right'
x[facets[facet_positions]]=array([[[0., 0.],
[0., 2.]]])
region='top'
x[facets[facet_positions]]=array([[[2., 0.],
[2., 2.]]])
region='left'
x[facets[facet_positions]]=array([[[2., 2.],
[0., 2.]]])
where we see that the bottom facet is marked correctly, but that the right facet maps to what should be the left facet, top should be right, and left should be top.
Could anyone shed some light on this mysterious indexing?
The figure of the mesh above is created by converting the mesh to medit and read through meshio to convert to vtk, where the indexing is even stranger (goes from 2->5):
while converting the meshio to Medit and then to vtk, results in the following markers:
import meshio
mesh = meshio.read("tmp.mesh")
def create_mesh(mesh, cell_type, prune_z=False):
cells = mesh.get_cells_type(cell_type)
cell_data = mesh.get_cell_data("medit:ref", cell_type)
points = mesh.points[:, :2] if prune_z else mesh.points
out_mesh = meshio.Mesh(
points=points, cells={cell_type: cells}, cell_data={"name_to_read": [cell_data]}
)
return out_mesh
triangle_mesh = create_mesh(mesh, "triangle")
line_mesh = create_mesh(mesh, "line")
meshio.write("facets.vtk", line_mesh)
meshio.write("cells.vtk", triangle_mesh)