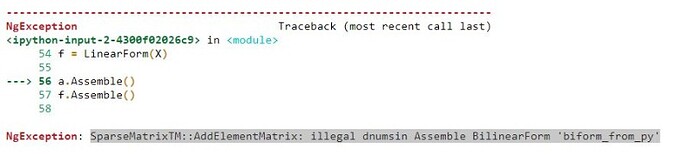
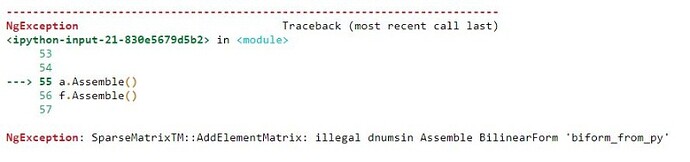
- You want to use “element_boundary” here. “skeleton” does not give you what you want.
“skeleton” terms are integrals over facets, but there are multiple volume elements attached to a facet.
The term
a += h*(p-pbar)*(q-qbar)*dx(skeleton=True)
is not well defined. Which of the volume elements attached to a facet should provide “p” and “q”?
With “element_boundary”, it is well defined. Every facet is integrated over twice - once as part of the boundary of each volume element.
“skeleton” is usually used when you have jumps between volume elements, something like
a += h*(p-p.Other())*(q-q.Other())*dx(skeleton=True)
Here, it does not matter which of the volume elements of a facet provides “p” and which one “p.Other()”.
To make this clear, your term, using “skeleton”, would be something like this:
a += h * ( (p-pbar)*(q-qbar) + (p.Other()-pbar)*(q.Other()-qbar) ) * dx(skeleton=True)
For “skeleton” to work, though, you have to set “dgjumps=True” in the FESpace:
X = FESpace([V, Vbar, Vbar, Q, Qbar], dgjumps=True)
Forgetting this, and using “skeleton” integrals leads to the illegal dnums error you encountered.
-
See the replies to this post from last week as for why your problem is not well posed if we use Dirichlet conditions everywhere and how to fix this issue.
-
There is a typo:
(u, ubar1, ubar2, p, pbar) = X.TrialFunction()
(v, vbar1, vbar2, q, qbar) = X.TrialFunction()
should be
(u, ubar1, ubar2, p, pbar) = X.TrialFunction()
(v, vbar1, vbar2, q, qbar) = X.TestFunction()
You can do
a.Assemble()
print(a.mat)
and look at the matrix entries as a first sanity check. In this case, ALL entries are 0, which leads us to suspect something is off with trial/test functions.
- Your formulation is wrong
bh_dx = - p*div(v) + q*div(u)
bh_ds = (v-vbar)*n*pbar - (u-ubar)*n*qbar,
should, I think, be
bh_dx = -p*div(v) - q*div(u)
bh_ds = v*n*pbar + u*n*qbar
You should multiply the divergence criterium by -1, or define p as -p such that you get a symmetric matrix. This is not strictly speaking necessary, but good practice.
The term (v-vbar)npbar is, I believe, wrong. I think it should be a bnpbar
Best,
Lukas
Attachment: sto.py