Hi, this is a follow-up on my previous post (the same model, but different problem). I am trying to implement a system of coupled PDEs, but I am not sure if I’m doing it correctly.
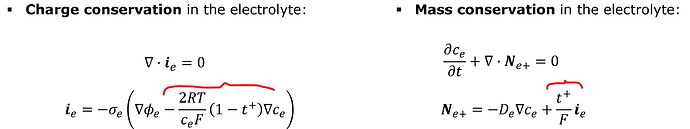
Let’s first look at the problem without the coupling:
This one is straightforward to implement based on the available examples:
fes_ele_v = H1(mesh, order=2, definedon="ele", dirichlet="left_ele|right_ele|top_ele|bottom_ele")
fes_ele_c = H1(mesh, order=2, definedon="ele")
fes_ele = fes_ele_v * fes_ele_c
(u_phie, u_ce), (v_phie, v_ce) = fes_ele.TnT()
gfe = GridFunction(fes_ele)
gf_phie, gf_ce = gfe.components
a_e = BilinearForm(fes_ele, symmetric=False)
a_e += D_e_eff * grad(u_ce) * grad(v_ce) * dx(definedon=mesh.Materials("ele"))
a_e += sigma_e_eff * grad(u_phie) * grad(v_phie) * dx(definedon=mesh.Materials("ele"))
a_e.Assemble()
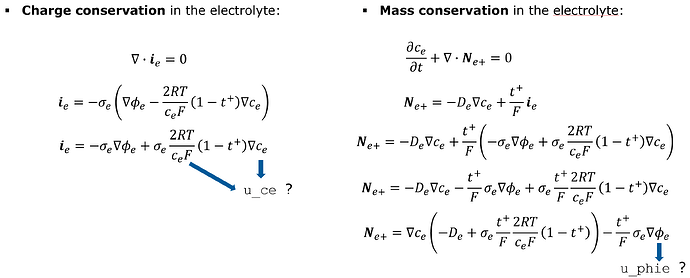
However, the full model contains the coupling between the electric and diffusion equations:
There I’m not sure if c_e from the left equations should be a test function u_ce, or a grid funnction gf_ce. And the similar question for phi_e that will appear on the right side when i_e is expanded
Option 1: We expand the equations and use test functions:
And the bilinear form looks like this:
a_e = BilinearForm(fes_ele, symmetric=False)
a_e += (-D_e_eff + 2 * sigma_e_eff * t_plus * (1 - t_plus) / F / FoverRT) * grad(u_ce) * grad(v_ce) * dx(definedon=mesh.Materials("ele"))
a_e += -t_plus / F * sigma_e_eff * grad(u_phie) * grad(v_phie) * dx(definedon=mesh.Materials("ele"))
a_e += sigma_e_eff * grad(u_phie) * grad(v_phie) * dx(definedon=mesh.Materials("ele"))
a_e.Assemble()
a_e += -sigma_e_eff * 2 / FoverRT / u_ce * (1 - t_plus) * grad(u_ce) * grad(v_phie) * dx(definedon=mesh.Materials("ele"))
Option 2: use c_e and phi_e in the respective charge and mass equations as a posteriori grid functions from the previous timestep, and calculate divergences directly:
ie_flux = GridFunction(HDiv(mesh, order=2, definedon="ele"))
ce_flux = GridFunction(HDiv(mesh, order=2, definedon="ele"))
a_e = BilinearForm(fes_ele, symmetric=False)
a_e += D_e_eff * grad(u_ce) * grad(v_ce) * dx(definedon=mesh.Materials("ele"))
a_e += -t_plus / F * div(ie_flux) * dx(definedon=mesh.Materials("ele"))
a_e += sigma_e_eff * grad(u_phie) * grad(v_phie) * dx(definedon=mesh.Materials("ele"))
a_e.Assemble()
a_e += -sigma_e_eff * 2 / FoverRT / gf_ce * (1 - t_plus) * div(ce_flux) * dx(definedon=mesh.Materials("ele"))
Fluxes are calculated in every time increment, something like this:
ie = -sigma_e_eff * (grad(gf_phi) - 2 / FoverRT / c * 0.5 * grad(c))
ie_flux.Set(ie)
ce_flux.Set(gf_ce)